BIOPRED: A Machine Learning-Based Web Application CDD
Client or Institution: Academic Thesis, Yildiz Technical University (YTU)
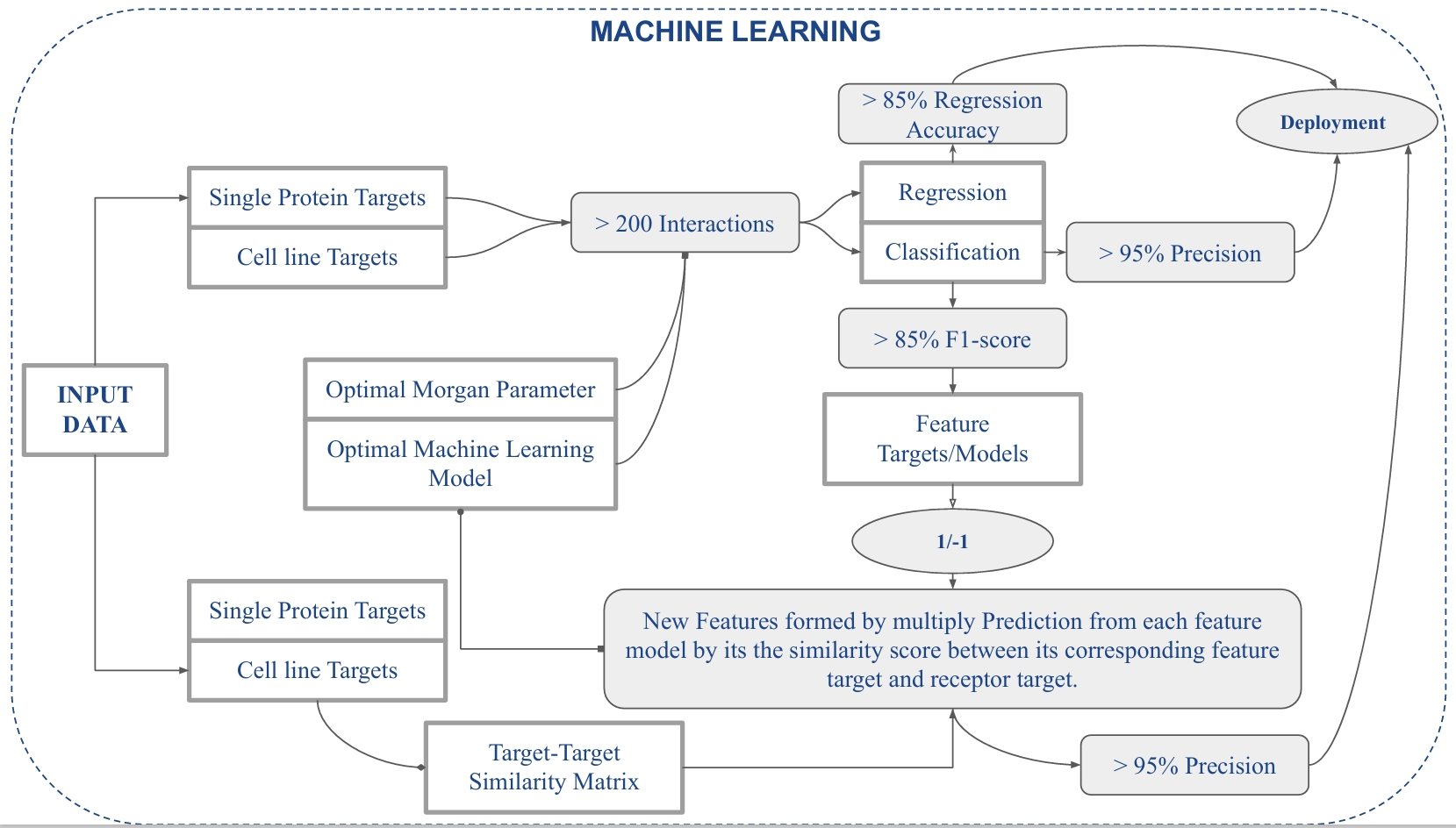
- Project Goal: Build predictive machine learning models to estimate bioactivity between small molecules and protein targets using ChEMBL data, and expose the functionality via a user-friendly web app for drug repurposing and molecular docking support
- Tools Used: Python, Scikit-learn, Streamlit, RDKit, Pandas, NumPy
- Duration: 6 months
- Outcome: Developed a Streamlit-powered platform capable of both regression and classification-based activity prediction, optimized through experimental benchmarking, with precision up to 95% in some tasks
- Data Source: ChEMBL (bioactivity), supplemented with local database formats
Built on ChEMBL Data, Scikit-Learn Models, and Streamlit
Abstract
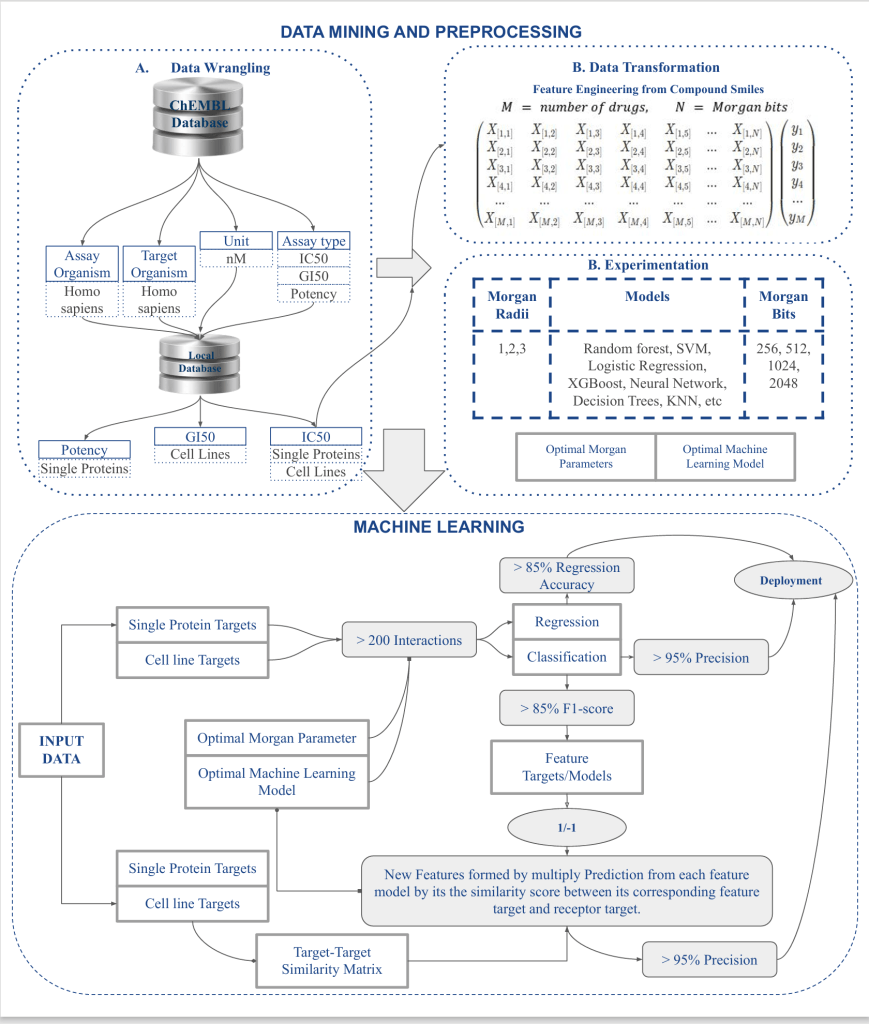
BIOPRED is a research-driven bioinformatics platform that uses machine learning algorithms to predict drug-target interactions, assist in drug repurposing, and support molecular docking workflows. By transforming molecular SMILES into numerical fingerprints using Morgan bits, and feeding them into supervised learning models, BIOPRED enables predictions of continuous bioactivity values (IC50, GI50, potency) or binary activity classification (active/inactive).
The project focuses on two prediction scenarios:
- Single-protein targeting using regression
- Activity classification for known compounds
The platform was designed to be scalable, reproducible, and interactive—enabling researchers to explore bioactivity predictions using a simple web interface.
🧠 Background & Problem Statement
Predicting the biological activity of chemical compounds is a central task in drug discovery. Traditional approaches are slow and resource-intensive. Modern cheminformatics offers a faster route using ML algorithms trained on public bioactivity datasets like ChEMBL.
However, challenges include:
- Feature engineering from chemical structures
- Choosing the right fingerprinting technique (Morgan bits, radius)
- Benchmarking the best models across regression and classification pipelines
- Balancing predictive accuracy with computational efficiency for real-world use
BIOPRED addresses all these via a well-structured ML pipeline and a user-accessible interface.
🗂 Dataset & Preprocessing
📌 Source:
- ChEMBL Database – extracted SMILES strings, IC50, GI50, assay type, and organism data
- Focused on Homo sapiens targets with >200 compound-protein interaction records
🧪 Target Types:
- Single Protein Targets
- Cell Line Targets
🛠 Preprocessing Workflow:
- SMILES strings converted to Morgan fingerprints using RDKit
- Feature sets: 256, 512, 1024, 2048 bits
- Morgan radii: 1, 2, and 3
- Labels: Continuous IC50 (for regression), Binary classification (1 for active, -1 for inactive)
⚙️ Machine Learning Pipeline
🔁 Algorithms Used
- Regression: SVR, LinearSVR, Random Forest, Lasso, Decision Tree, MLP, XGBoost
- Classification: Random Forest, XGBoost (top performers), Logistic Regression, SVM
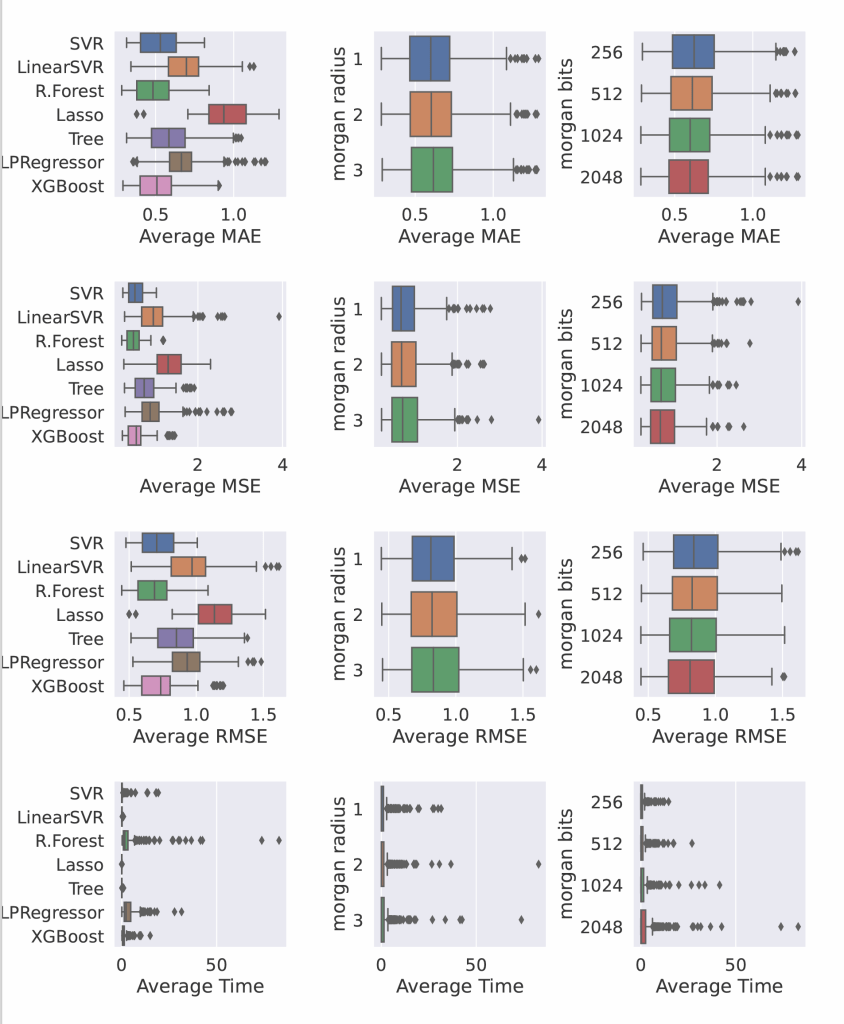
📈 Model Evaluation
Regression Metrics:
- MAE, MSE, RMSE
- Top Regression Accuracy: 85%
- Best Models: XGBoost, MLPRegressor
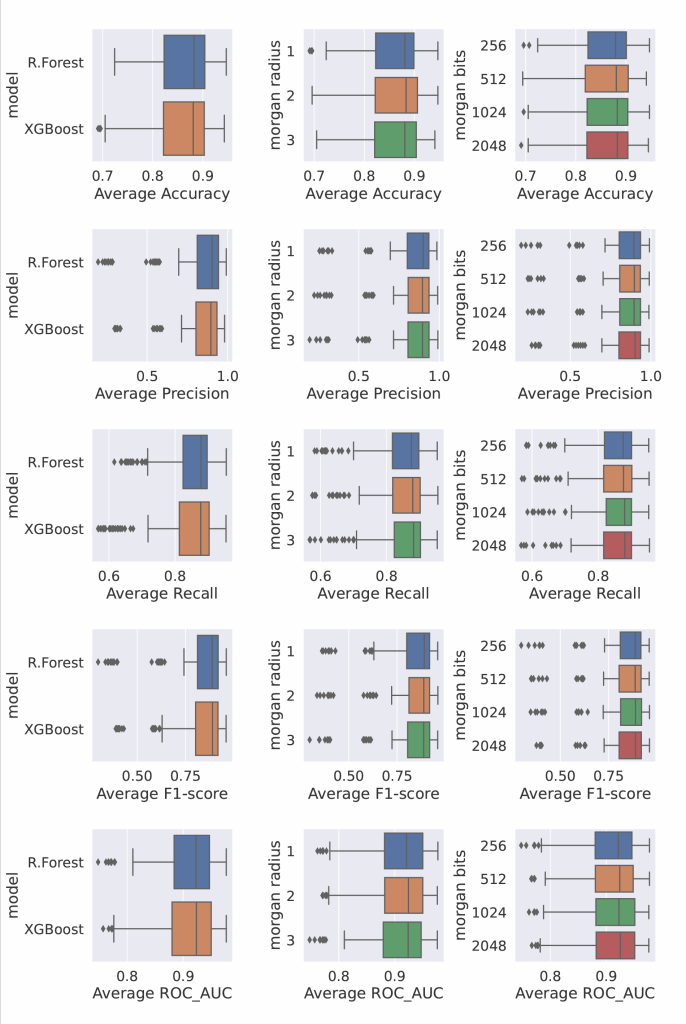
Classification Metrics:
- Accuracy, Precision, Recall, F1-score, ROC AUC
- Top Classification Precision: >95%
- Best Models: Random Forest, XGBoost
💡 Model Optimization Insights
- Morgan bit length of 1024–2048 and radius of 2 or 3 gave best trade-off between performance and computation
- XGBoost consistently outperformed other models on both tasks
- Model performance was highly sensitive to class balance and compound-target diversity
- Feature-target similarity scoring helped boost model predictions in novel target tasks
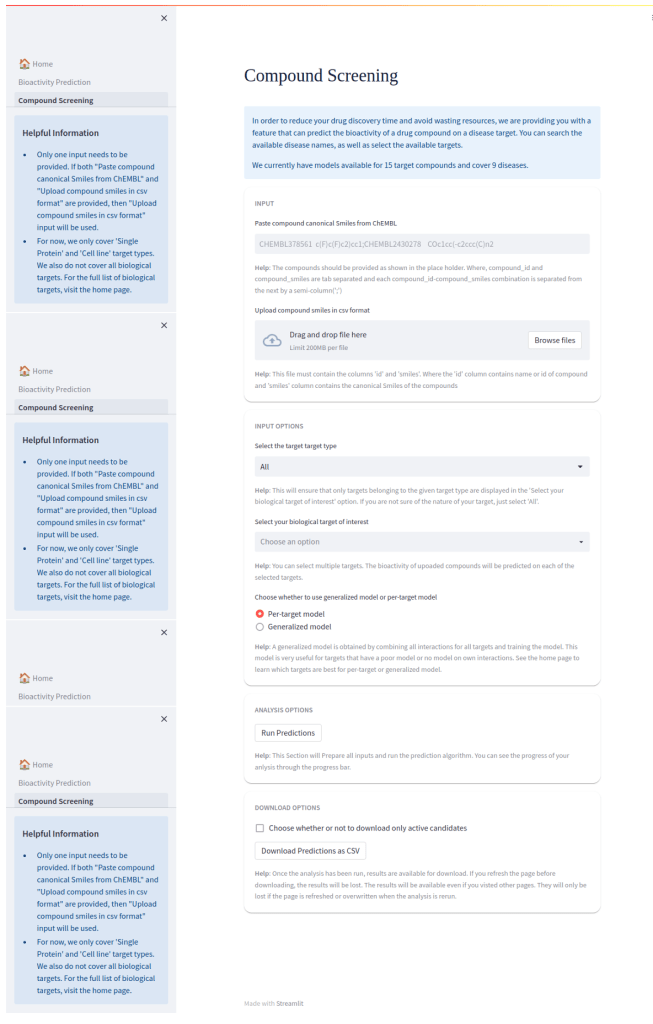
🌐 Web Application (BIOPRED)
The BIOPRED Streamlit web app allows users to:
- Input a SMILES string
- Choose task type (regression/classification)
- Select protein target or assay setting
- Get real-time predictions on:
- Activity values (IC50, GI50)
- Activity class (active/inactive)
- Visualize results with confidence levels
📈 Outcome
- Trained and evaluated dozens of ML model variants on ChEMBL interaction data
- Delivered prediction scores with high precision and recall
- Deployed results in a fully functional Streamlit app with user input and result feedback
- Contributed code and documentation to GitHub for transparency and reproducibility
- Validated performance via experimental result charts and precision metrics
- Positioned as a pipeline-ready tool for drug discovery workflows
💬 Discussion
BIOPRED combines strong ML architecture with practical biomedical relevance. It tackles a core challenge in cheminformatics: translating SMILES strings into actionable predictions. The project also demonstrates how a thesis project can integrate deep technical components with real-world usability through interface design and reproducibility.
The success of this project lies in:
- Balancing performance with interpretability
- Making the tool accessible via the web
- Adhering to academic rigor in model reporting and experimentation
📣 Call to Action
Have a list of SMILES strings or target proteins and want to test their bioactivity?
Interested in building your own prediction tool?
💬 Start a conversation .

Leave a comment