Wrapping Bioinformatics Tools into a User-Friendly Web Platform
Simplify and democratize access to advanced bioinformatics tools by transforming complex command-line operations into an intuitive, user-friendly web interface. This cloud-enabled platform empowers researchers, students, and professionals to run cutting-edge analyses without needing deep computational expertise, accelerating discovery and fostering broader engagement in bioinformatics across various disciplines and backgrounds.
- Client or Institution: Bioinfopipe Ltd (Startup Project)
- Tools Used: Docker, Command Line, GitHub, Nextflow (used not developed), Tool Docs
- Duration: 4 months
- Outcome: Successfully wrapped 30+ tools and 300+ tool subcommands; built robust documentation; developed strong debugging and integration experience
Project URL: https://www.bioinfopipe.com/
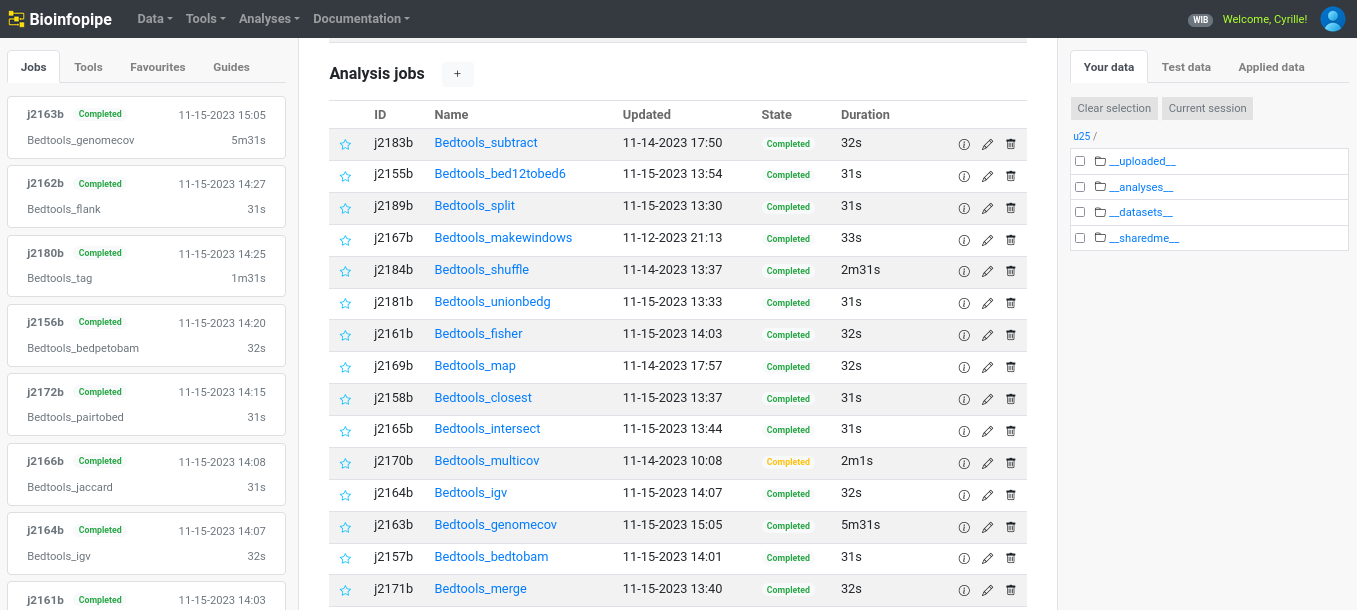
Transforming Command-Line Complexity into One-Click Simplicity
Abstract
In this project for Bioinfopipe Ltd, I helped build a web-based platform that wraps complex bioinformatics tools into user-friendly buttons. By abstracting command-line instructions and integrating Dockerized tools from AWS ECR and BioContainers, we created a seamless, cloud-supported solution that democratizes access to bioinformatics workflows. This post details how I navigated tool wrapping, documentation gaps, testing, and rapid scaling of tool integration.
Background & Problem Statement
Bioinformatics tools are powerful—but often inaccessible to non-technical users due to their complex command-line interfaces and steep learning curves. Bioinfopipe Ltd set out to change that by launching a platform that turns these tools into simple UI buttons backed by scalable AWS cloud infrastructure.
My role was to wrap tools, test subcommands, ensure documentation clarity, and provide consistent user-level access without compromising tool integrity. The challenge: many tools lacked usable instructions or test datasets, requiring creativity, patience, and strong documentation literacy.
Dataset & Resources
There was no unified dataset; instead, each tool required:
- Sample datasets sourced or generated synthetically
- Reference command-line instructions from GitHub repos or tool documentation
- Internal test pipelines to verify tool behavior after wrapping
All tools were tested independently using synthetic or published example data to confirm consistency and reproducibility.
Approach & Methodology
Here’s how the wrapping process worked:
🔹 Tool Containerization
- Imported Docker images from BioContainers using AWS ECR.
- Ensured compatibility with Bioinfopipe’s architecture.
🔹 Wrapping Workflow
- Used Nextflow to wrap tool commands and subcommands.
- Created modular wrappers for each tool, specifying parameters and tags.
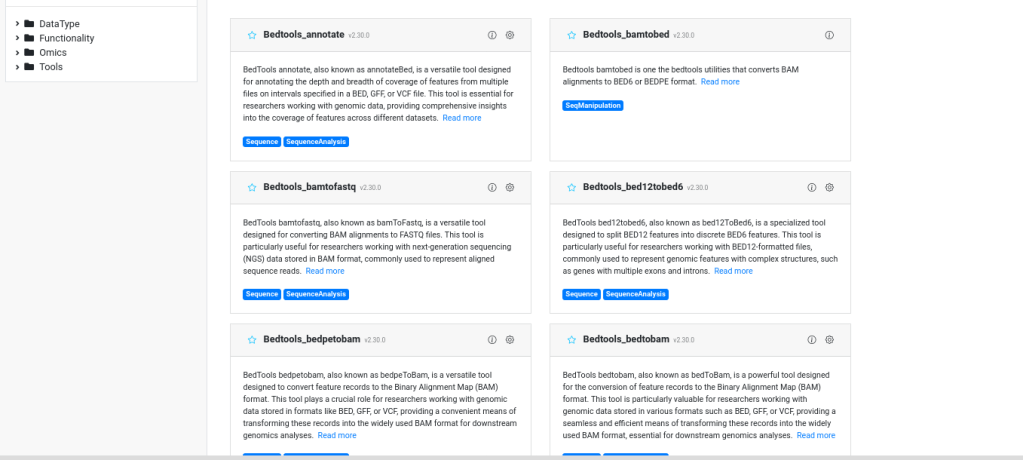
🔹 Documentation Creation
- Each wrapped tool came with a custom documentation file including:
- Tool description
- Input/output format
- Parameter definitions
- Sample usage instructions
🔹 Testing
- Conducted test runs on dummy or real datasets.
- Manually verified tool performance and resolved path, syntax, or resource issues.
Key Results
- 30+ Tools Wrapped — including multiple categories (e.g., QC, Alignment, Variant Calling, Annotation)
- 200+ Subcommands Integrated — offering fine-grained control from simple UI elements
- Complete Documentation — allowing even non-technical users to explore tools confidently
- Deep Debugging Skills Acquired — from error tracing to Docker image exploration
- Scalable Setup — used the pre-existing AWS & Nextflow integration without modifying infrastructure
Lessons Learned / Innovations
What Worked:
- Gaining mastery over three tools made wrapping subsequent ones dramatically faster.
- Reading code documentation effectively became a superpower—I learned to find what I needed quickly and decode undocumented behavior through trial runs and literature review.
Challenges:
- Some tools lacked any usable instructions or real test data.
- Resolved gaps using synthetic test data and deep dives into academic papers or GitHub issues.
Key Takeaway:
“Don’t assume you know. Don’t fear what you don’t.”
This mindset helped me grow rapidly during this project.
Discussion
This project wasn’t just about technical wrapping—it was about making bioinformatics accessible. It showed how much of science’s complexity is hidden behind interfaces. By transforming command-line burdens into clickable workflows, we help expand the reach of bioinformatics to researchers, students, and even clinicians.
The final system not only runs seamlessly via cloud resources but provides a robust framework for wrapping any future tool.
Though I didn’t set up the AWS-Nextflow pipeline myself, working within this ecosystem gave me real-world exposure to cloud-based workflow execution and scalable design.
This was a client-based project, and future steps remain confidential. But if you’re working on a bioinformatics platform or need tool integration support, feel free to start a conversation or view more projects.
Final Thoughts & Future Directions
Projects like this aren’t just technical exercises—they are steps toward democratizing science. I’d love to see more open bioinformatics platforms embrace this model. Moving forward, I aim to apply this experience to future client work involving cloud workflows, tool wrapping, or user interface design for scientific tools.

Leave a comment