A collection of projects, collaborations, and research work where I’ve applied bioinformatics, data science, and coding to solve real problems and support scientific discovery.
-
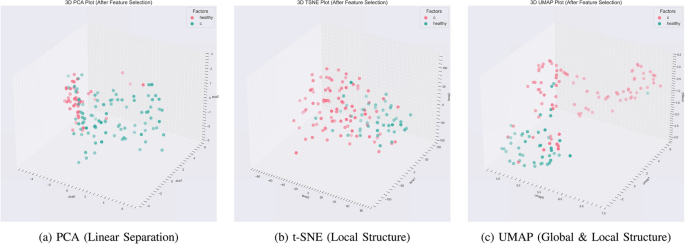
Ensemble Feature Selection for Robust Multi-Omics Biomarker Discovery
Ensemble Feature Selection for Robust Multi-Omics Biomarker Discovery A Systems-Level Approach to Biomarker Discovery Abstract Biomarker discovery often suffers from instability — different feature selection algorithms produce different gene lists, reducing reproducibility and clinical trust. This project addressed that challenge by designing and implementing a comprehensive ensemble feature selection framework that integrates multiple ranking strategies…
-
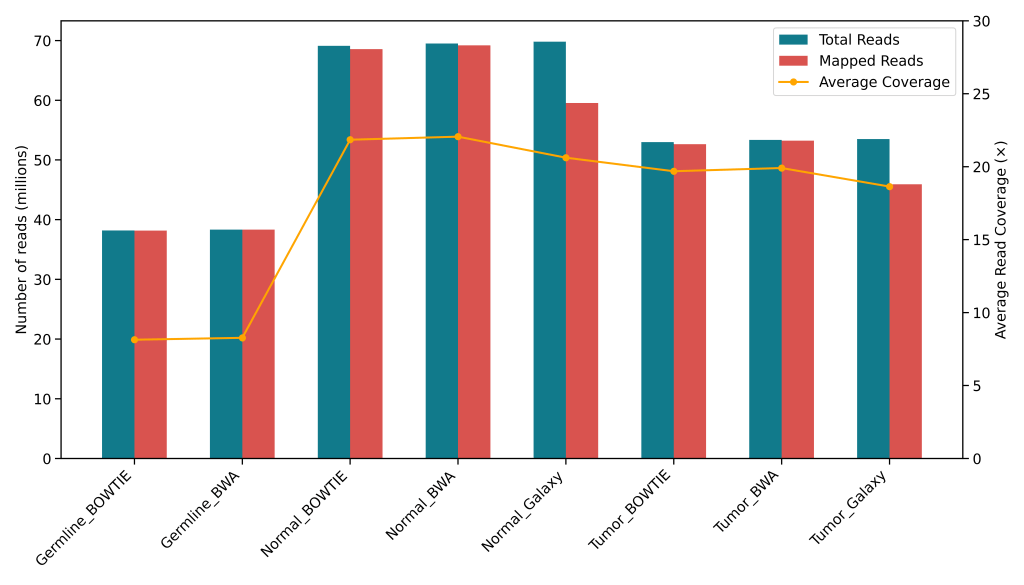
Benchmarking Germline and Somatic Variant Calling Pipelines Using CoSAP and Galaxy
Benchmarking Germline and Somatic Variant Calling Pipelines Using CoSAP and Galaxy A Comparative Study with 9 Pipelines on Real Exome Data Abstract This project evaluated nine variant calling pipelines using whole exome sequencing (WES) data. It involved combinations of two mappers (BWA, Bowtie2) and four variant callers (DeepVariant, HaplotypeCaller, Strelka, SomaticSniper) across both germline and…
-
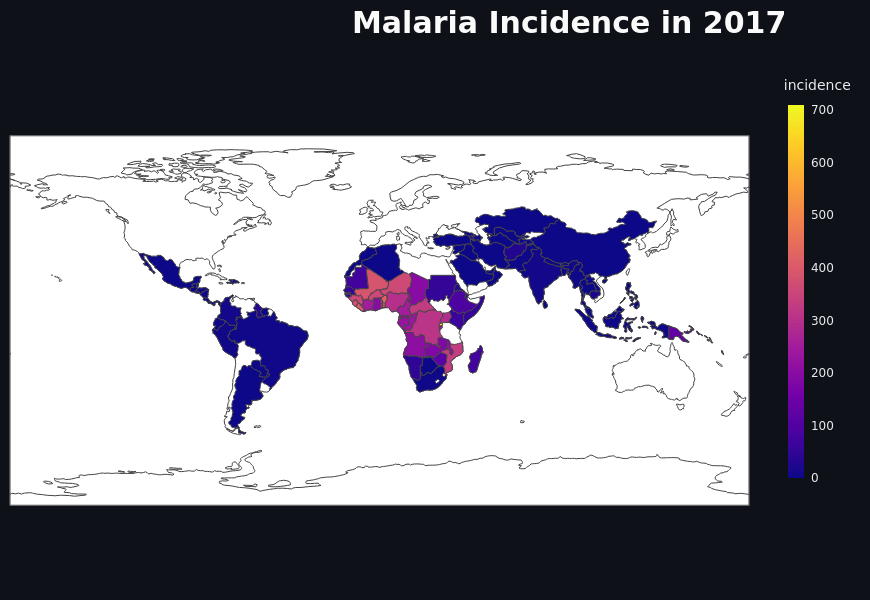
Predicting Malaria Incidence from Climate Data Using Machine Learning
The project aimed to predict malaria incidence using climate and geographical data through machine learning, deploying a Streamlit web app for visualization across 98+ countries. With data from WHO and others, the CatBoost model achieved a 96.7% correlation. It provides easily accessible insights for researchers and policymakers, addressing malaria’s global health challenge.
-

AutomatedMLPack: A Python Package for End-to-End Automated Machine Learning
AutomatedMLPack is a Python package designed for streamlined automated machine learning workflows, enabling efficient data ingestion, model training, and evaluation through a command-line interface. It supports classification and regression tasks, offers flexible feature selection, and provides visualizations and evaluation reports, significantly enhancing productivity in machine learning projects.
-
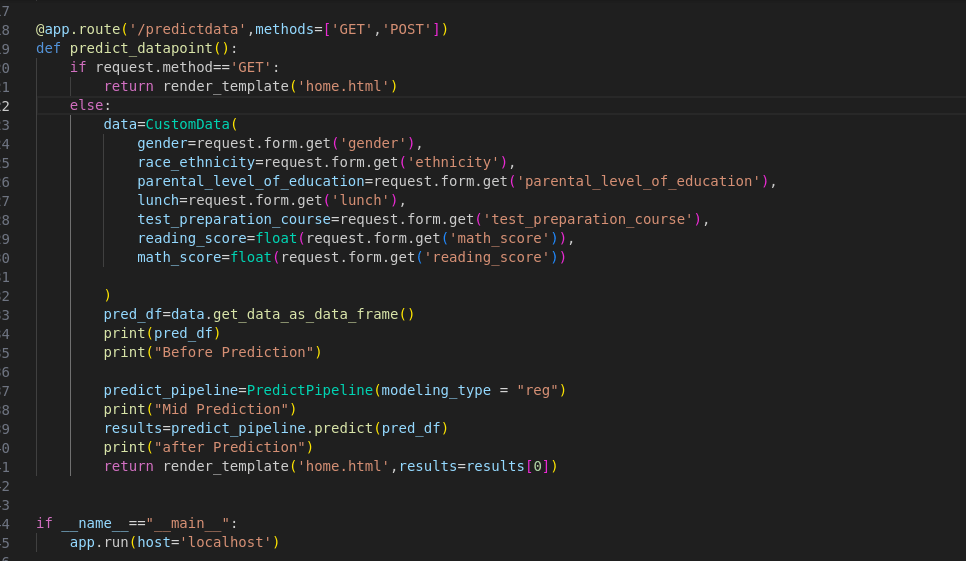
Building Scalable ML Applications: A Practical Approach
The project involved developing a comprehensive machine learning pipeline for classification and regression tasks, culminating in a Flask-based web application deployed on Azure. It features automated deployment via GitHub Actions, ensuring a user-friendly interface for real-time predictions. Key achievements include a modular pipeline and seamless integration, enhancing accessibility in ML applications.
-
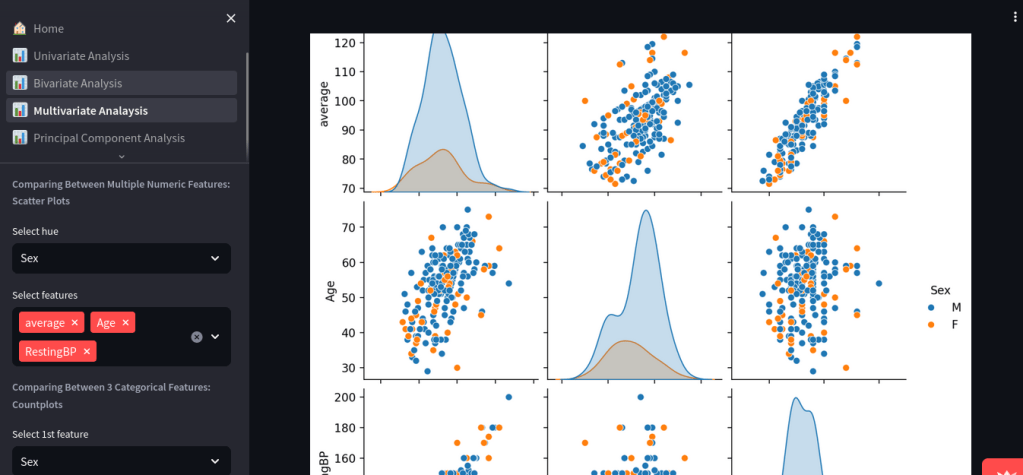
Dynamic Exploratory Data Analysis with Streamlit
The Dynamic Exploratory Data Analysis app simplifies EDA for users of all skill levels by allowing CSV uploads and generating insightful visualizations. Developed with Streamlit, it automates data type detection and offers various analysis modules. Key features include univariate, bivariate, and multivariate visualizations, making data exploration accessible and effective.
-
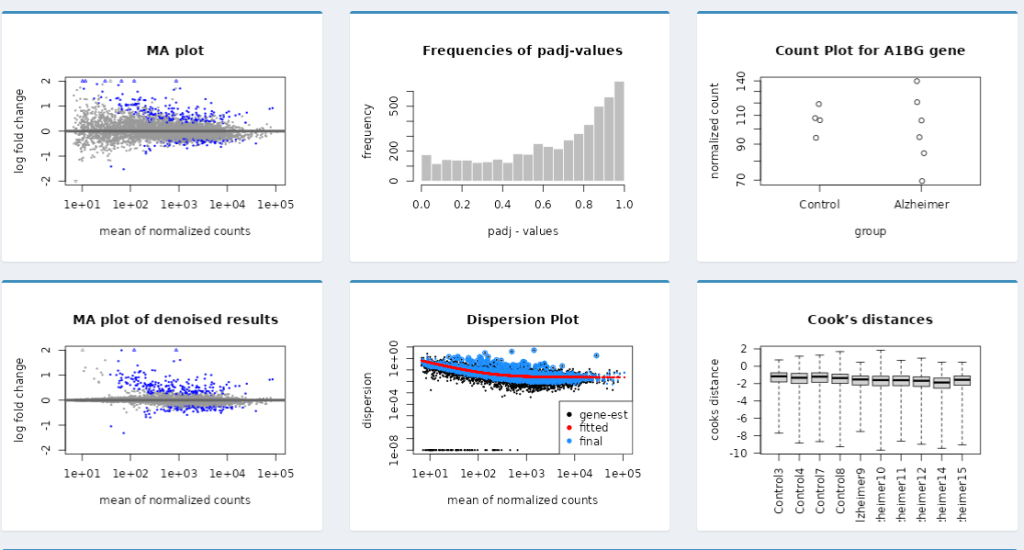
Dynamic Shiny Dashboard for the Visualization of DESeq2 Results
The project developed an interactive Dynamic Shiny Dashboard to visualize DESeq2 differential expression results, allowing users to explore data without R programming skills. It features various visualizations such as volcano plots and heatmaps, facilitating effective communication of results while supporting collaborative efforts in bioinformatics. The project lasted two weeks.
-
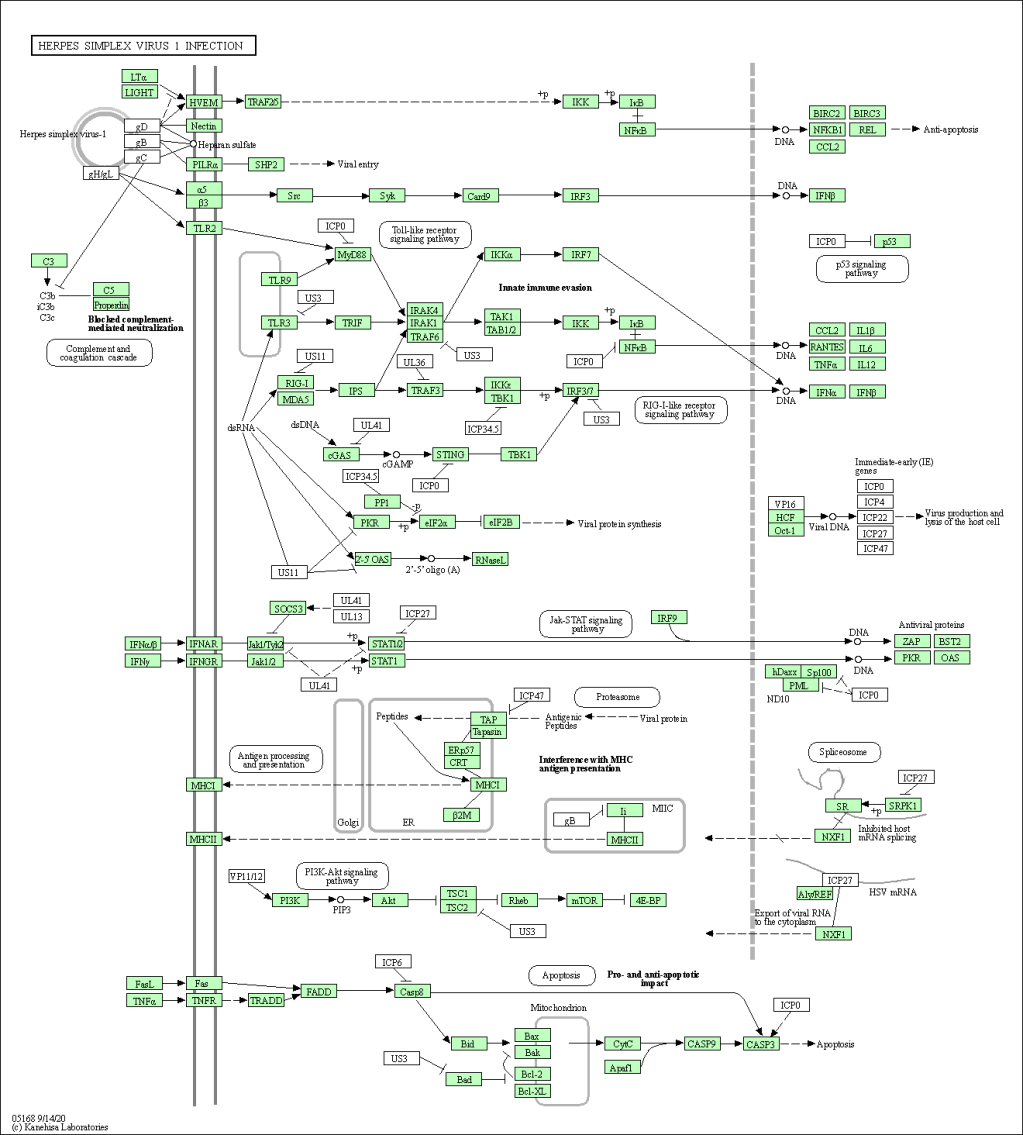
Gene Set Enrichment and KEGG Pathway Analysis Using ClusterProfiler
The project utilized KEGG-based gene set enrichment analysis from DESeq2 results to visualize biological pathway alterations in Alzheimer’s disease. Using R and ClusterProfiler, enriched pathways were identified and visualized, revealing significant immune and neurodegenerative responses. The findings could inform future research and biomarker discovery in Alzheimer’s.
-
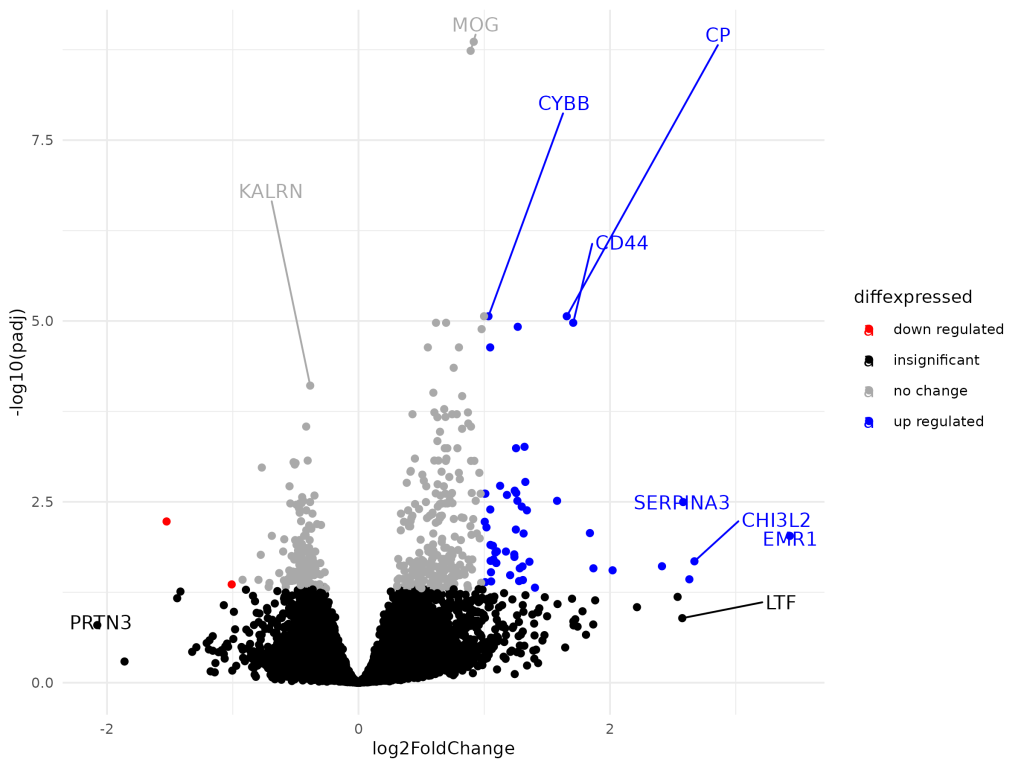
Differential Gene Expression Analysis in Alzheimer’s Disease
This project conducts differential gene expression analysis on Alzheimer’s disease using RNA-Seq data from the GSE53697 dataset, identifying differentially expressed genes (DEGs) via DESeq2 in R. It emphasizes preprocessing, outlier removal, and visualization through a Shiny app, facilitating interactive exploration of results, enhancing understanding of gene expression changes.
-
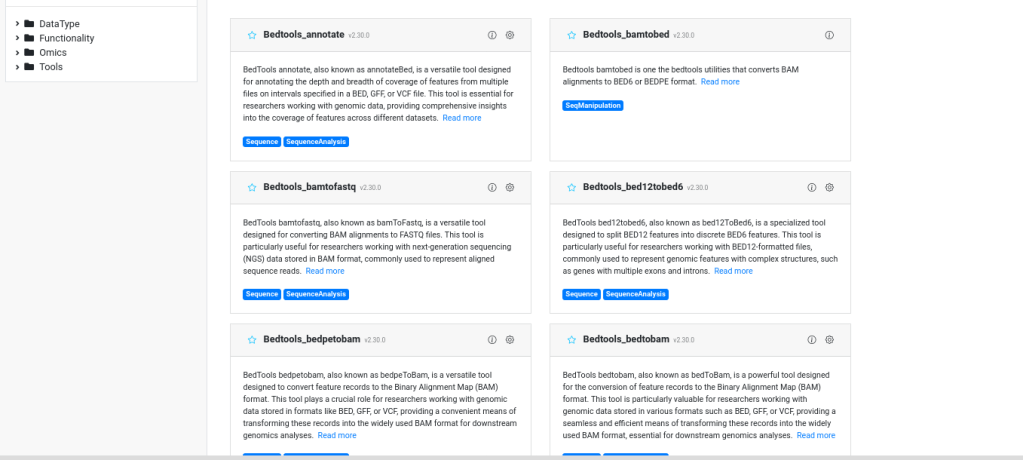
Wrapping Bioinformatics Tools into a User-Friendly Web Platform
The project aimed to simplify access to complex bioinformatics tools by creating a user-friendly web platform for Bioinfopipe Ltd. Over four months, over 300 tools were wrapped into intuitive interfaces, enhancing usability for non-technical users. The project resulted in comprehensive documentation and a scalable solution on AWS, democratizing bioinformatics.

