Tag: Bioinformatics
-
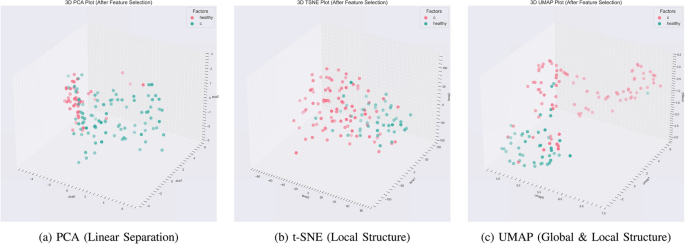
Ensemble Feature Selection for Robust Multi-Omics Biomarker Discovery
Ensemble Feature Selection for Robust Multi-Omics Biomarker Discovery A Systems-Level Approach to Biomarker Discovery Abstract Biomarker discovery often suffers from instability — different feature selection algorithms produce different gene lists, reducing reproducibility and clinical trust. This project addressed that challenge by designing and implementing a comprehensive ensemble feature selection framework that integrates multiple ranking strategies…
-
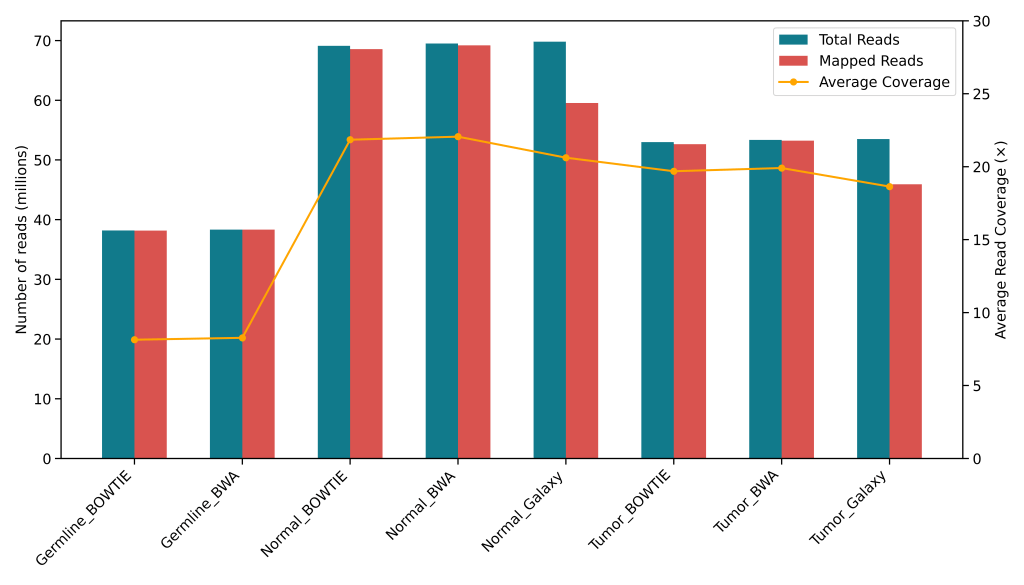
Benchmarking Germline and Somatic Variant Calling Pipelines Using CoSAP and Galaxy
Benchmarking Germline and Somatic Variant Calling Pipelines Using CoSAP and Galaxy A Comparative Study with 9 Pipelines on Real Exome Data Abstract This project evaluated nine variant calling pipelines using whole exome sequencing (WES) data. It involved combinations of two mappers (BWA, Bowtie2) and four variant callers (DeepVariant, HaplotypeCaller, Strelka, SomaticSniper) across both germline and…
-

Getting Started with Trimmomatic for Illumina Sequencing
Trimmomatic is a vital tool for preprocessing Illumina NGS data, addressing issues like adapter contamination and low-quality bases. It offers various quality filtering methods and supports paired-end reads. As an open-source tool, it enhances mapping accuracy and is essential for accurate downstream analyses in genomics research.
-
Quality Control with MultiQC: A Complete Guide
MultiQC is a bioinformatics tool that consolidates results from multiple samples into a single HTML report, streamlining data analysis from high-throughput sequencing. It supports over 100 tools, providing summary statistics and visualizations. This automation enhances quality control, saves time, and minimizes errors, making it ideal for various genomic workflows.
-
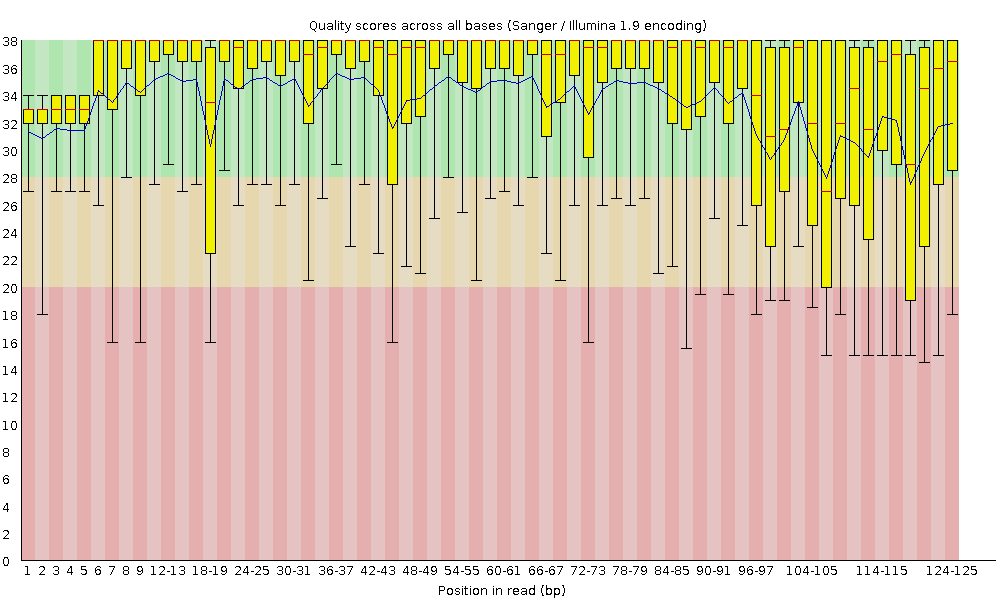
Quality Control of High-Volume Sequencing Data with FastQC: A Complete Guide
High-throughput sequencing technologies have transformed genomics by enabling massive data generation in record time. But with large volumes of data comes the responsibility of ensuring its quality. This is where FastQC steps in—offering a robust quality control solution tailored for sequencing data. 🔍 What is FastQC? FastQC is a Java-based application designed to analyze sequencing…
-

Exploring Essential Data Types and Formats in Bioinformatics: Origins and Applications
Bioinformatics merges biology and computational science to analyze complex biological data, vital for genomics, transcriptomics, and proteomics. Understanding data types like sequencing and file formats is essential for effective analysis. Each format addresses specific needs, enhancing data management and interpretation, crucial for impactful discoveries in life sciences and personalized medicine.
-
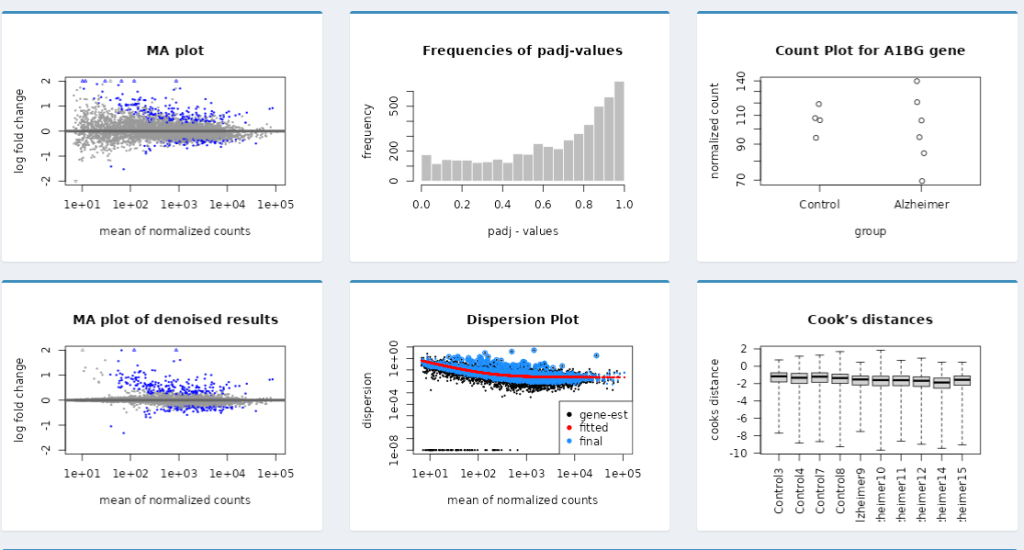
Dynamic Shiny Dashboard for the Visualization of DESeq2 Results
The project developed an interactive Dynamic Shiny Dashboard to visualize DESeq2 differential expression results, allowing users to explore data without R programming skills. It features various visualizations such as volcano plots and heatmaps, facilitating effective communication of results while supporting collaborative efforts in bioinformatics. The project lasted two weeks.
-
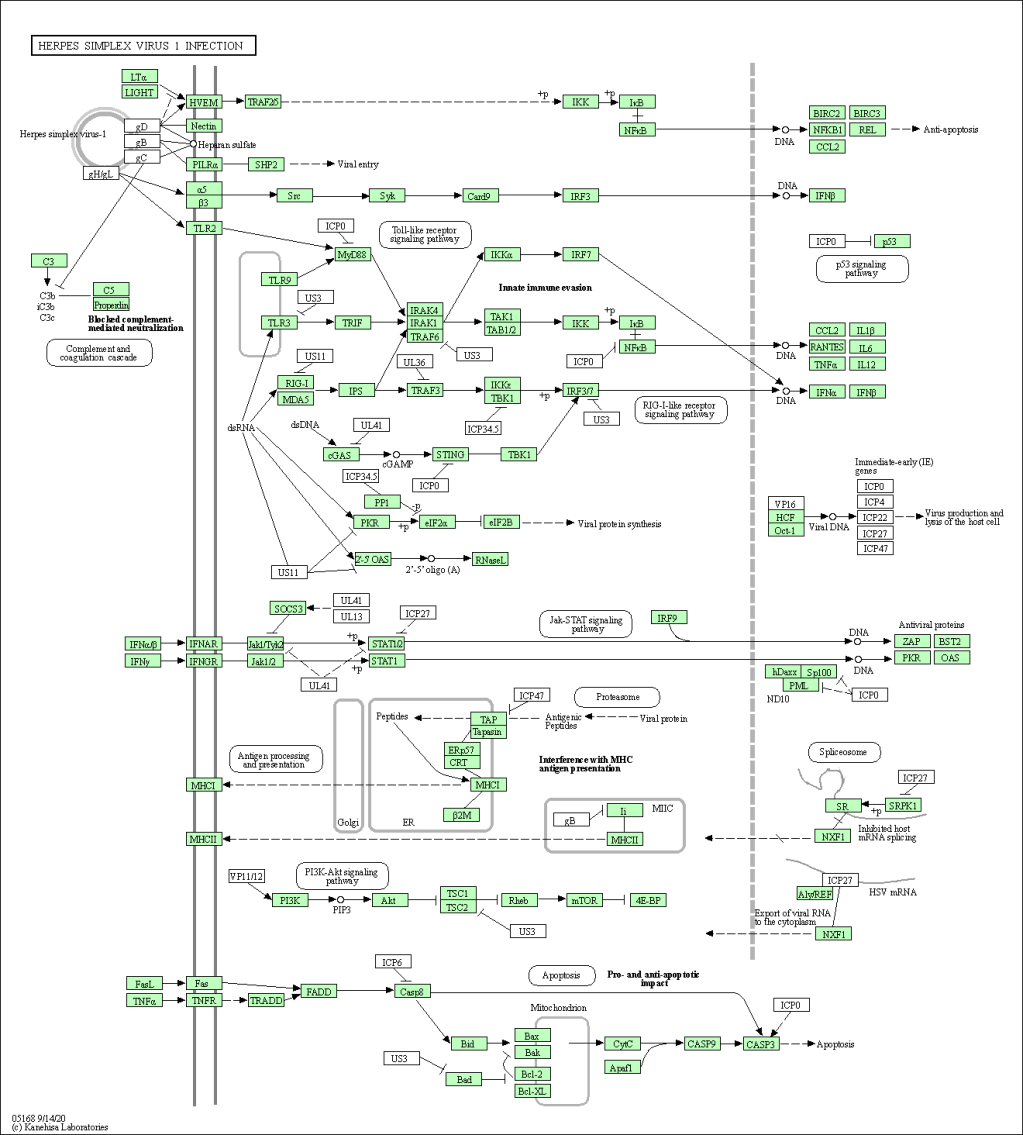
Gene Set Enrichment and KEGG Pathway Analysis Using ClusterProfiler
The project utilized KEGG-based gene set enrichment analysis from DESeq2 results to visualize biological pathway alterations in Alzheimer’s disease. Using R and ClusterProfiler, enriched pathways were identified and visualized, revealing significant immune and neurodegenerative responses. The findings could inform future research and biomarker discovery in Alzheimer’s.
-
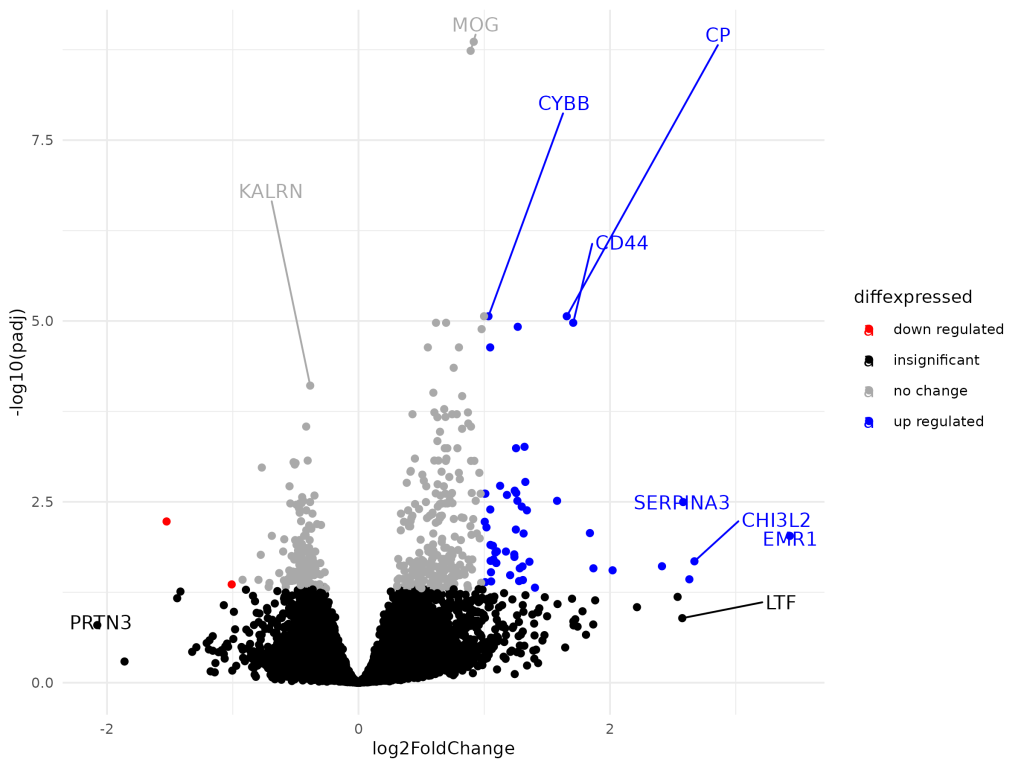
Differential Gene Expression Analysis in Alzheimer’s Disease
This project conducts differential gene expression analysis on Alzheimer’s disease using RNA-Seq data from the GSE53697 dataset, identifying differentially expressed genes (DEGs) via DESeq2 in R. It emphasizes preprocessing, outlier removal, and visualization through a Shiny app, facilitating interactive exploration of results, enhancing understanding of gene expression changes.
-
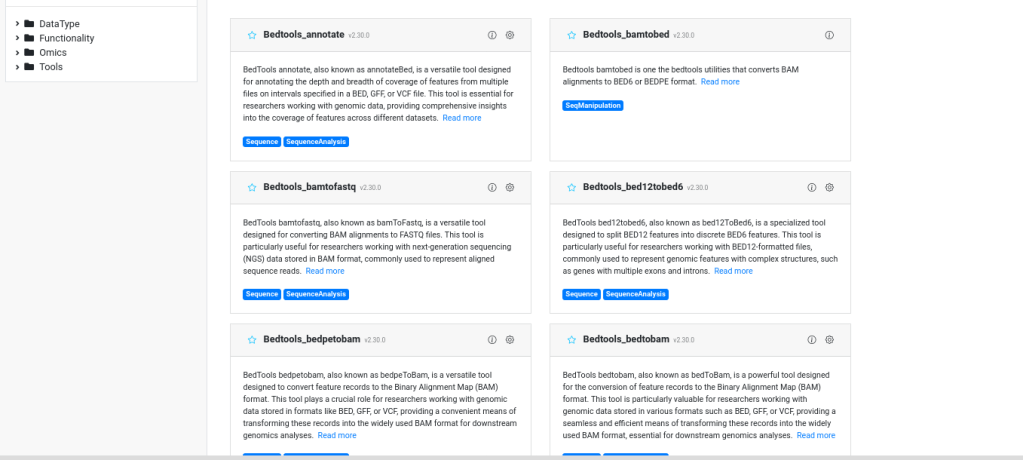
Wrapping Bioinformatics Tools into a User-Friendly Web Platform
The project aimed to simplify access to complex bioinformatics tools by creating a user-friendly web platform for Bioinfopipe Ltd. Over four months, over 300 tools were wrapped into intuitive interfaces, enhancing usability for non-technical users. The project resulted in comprehensive documentation and a scalable solution on AWS, democratizing bioinformatics.
